Rendering of peanut agglutinin-labeled proximal tubules (Figure 1), along with dialog boxes (left and center) used when making movies, and the graph-based color and opacity editor window (right).
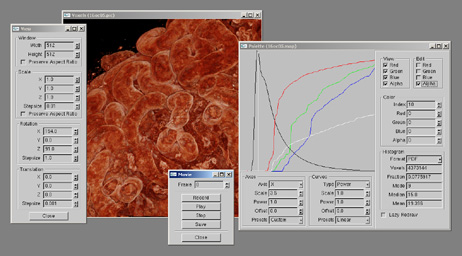
Figure 1 - Screen shot of Voxx
voxxajppaper.pdf - Journal article available in pdf format.
Abstract
Confocal and two-photon fluorescence microscopy have advanced the exploration
of complex, three-dimensional biological structures at submicron resolution.
We have developed a voxel-based three-dimensional imaging program (Voxx)
capable of near real-time rendering that runs on inexpensive personal computers.
This low-cost interactive 3D imaging system provides a powerful tool for analyzing
complex structures in cells and tissues and encourages a more thorough exploration
of complex biological image data.
Images and Movies
Full-size versions of the JPEG images can be displayed by clicking on the blue
"Figure xxx" label under the images. MPEG1 movies can be displayed by clicking
on the images or the blue "Movie xxx" labels under the images. Be aware that not
all web browsers have built-in support for MPEG1 movie playback. Microsoft Explorer
and Netscape Navigator on PCs commonly use Windows 98/2000's media player, and the
latest (free) version can be downloaded by clicking
HERE. Apple Macintosh users of Explorer
and Navigator need version 4.x or 5.x of the QuickTime player, and the latest
(free) version can be downloaded by clicking
HERE.
Rendering of peanut agglutinin-labeled proximal tubules (Figure 1), along with dialog boxes
(left and center) used when making movies, and the graph-based color and opacity
editor window (right).

Figure 1 - Screen shot of Voxx
Proximal tubules in the superficial cortex of newborn mouse labeled
with peanut agglutinin-rhodamine and imaged by two-photon microscopy (Figure 2).
In each case the field is 100 microns across.
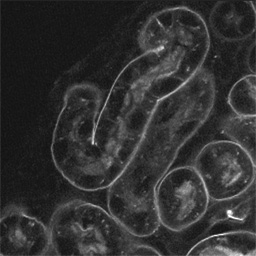
Figure 2A - A single optical section from the collected image volume.
Movie 2A -
Animation showing the sequential addition of 215 image planes as rendered using Voxx.
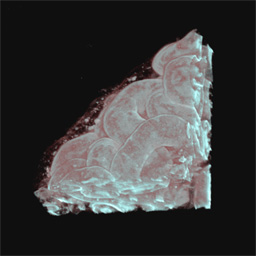
Figure 2B -
Entire volume as rendered using Voxx.
Movie 2B -
Animation showing the manipulation of the entire image volume.
More examples of microscopy volumes rendered using Voxx (Figure 3).

Figure 3A -
A rendered image volume of branching ureteric bud in a developing embryonic
kidney, labeled with peanut agglutinin-rhodamine and imaged using two-photon microscopy.
Movie 3A -
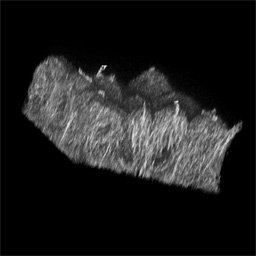
Figure 3B -
A rendered volume of polarized MDCK cells whose microtubules have been
labeled using anti-tubulin immunofluorescence, imaged by confocal microscopy.
Movie 3B -
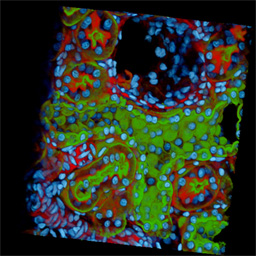
Figure 3C - Two-photon, triple labeling of rat kidney. Sections were labeled with the
nuclear stain DAPI (cyan), Texas Red phalloidin (red) to localize filamentous actin,
and a fluorescein conjugated lectin (green) from Dolichos biflouris to label the
collecting duct. The right side of the volume shows a partial section through a Bowmanís
capsule with the glomerulus missing. In the collecting duct (left), a checkerboard
staining pattern is produced by the localization of principal cells by the lectin (green).
Throughout the tubule, the nuclei from the intercalated cells are visible while those
from the principal cells are masked by lectin staining. In the upper left corner of the
volume, an arteriole staining intensely with the phallotoxin is seen. The actin rich
apical region of proximal tubules (interspersed throughout the volume) also stains
intensely with phalloidin.
Movie 3C -
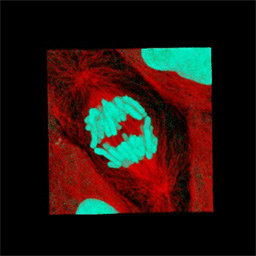
Figure 3D -
A confocal image volume of a dividing LLC-PK-1 cell, whose microtubules and
chromosomes have been labeled using anti-tubulin immunofluorescence and a DNA-binding
dye (TO-PRO-3), respectively. Movies 3A through 3D show these volumes being manipulated.
The field shown in panel A is 100 microns across, B is 40 microns across, C is 170 microns
across and D is 45 microns across.
Movie 3D -
Newborn mouse heart tissue, labeled with fluorescent wheat germ agglutinin,
and imaged by two-photon microscopy (Figure 4). Each field is 100 microns across.
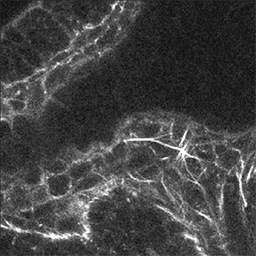
Figure 4A - A single focal plane.

Figure 4B -
The image volume rendered with a low opacity for low intensity voxels.
Movie 4B -
Shows the manipulation of this volume.

Figure 4C -
The same image volume rendered with high opacity for low intensity voxels.
Movie 4C -
Shows the manipulation of this volume.
Performance
We compared the rendering speeds of three systems (Figure 5). The rendering speed of the
image volume used for Figure 4 was recorded for the original 512 by 512 frame size and
for a 256 by 256 pixel subsection. In each case, the rendering speed was recorded after
loading a different number of optical sections. This test was conducted by running Voxx
on two PCs equipped with NVIDIA GeForce2 GTS and Quadro2 Pro graphics processors, while
a third system equipped with a VolumePro 500 voxel processor and 3Dlabs Oxygen GVX420
video board was running Revli.
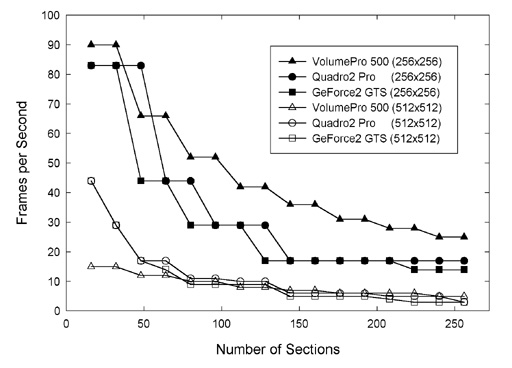
Figure 5 -
Rendering speed as a function of the size the image volume and of the
configuration of the host computer.